Faculty of Haide College achieved a significant progress in viral genome classification methods
Publisher:ZHAO
Release time:2025-03-26
Number of views:10
Recently, PhD candidate Zheng Kaiyang from the College of Marine Life Sciences and Assistant Professor Sun Jianhua from Haide College, served as co-first authors to develop a novel viral classification tool, VITAP (Viral Taxonomic Assignment Pipeline). Based on big data sequence alignment and graph theory methods, it provides cutting-edge technological support for research in viral metagenomics and viral ecology. Their achievements were published online on March 5 in Nature Communications, a sub-journal of the Nature publication. Professor Wang Min, Dean of Haide College and Vice Dean of the College of Marine Life Sciences, Professor Liang Yantao from the College of Marine Life Sciences, and Professor Andrew McMinn from the University of Tasmania, Australia, are the corresponding authors.
By combining sequence alignment and graph theory algorithms, VITAP enables highly accurate classification of both DNA and RNA viruses. It also provides confidence assessments for each taxonomic unit and can efficiently classify viral genome sequences as short as 1,000 base pairs across taxonomic levels from phylum to genus, achieving significantly improved viral annotation rates. Additionally, VITAP supports automatic updates based on the latest classification database from the International Committee on Taxonomy of Viruses (ICTV) and allows users to customize classification databases, offering enhanced user experience. (Figure 1).
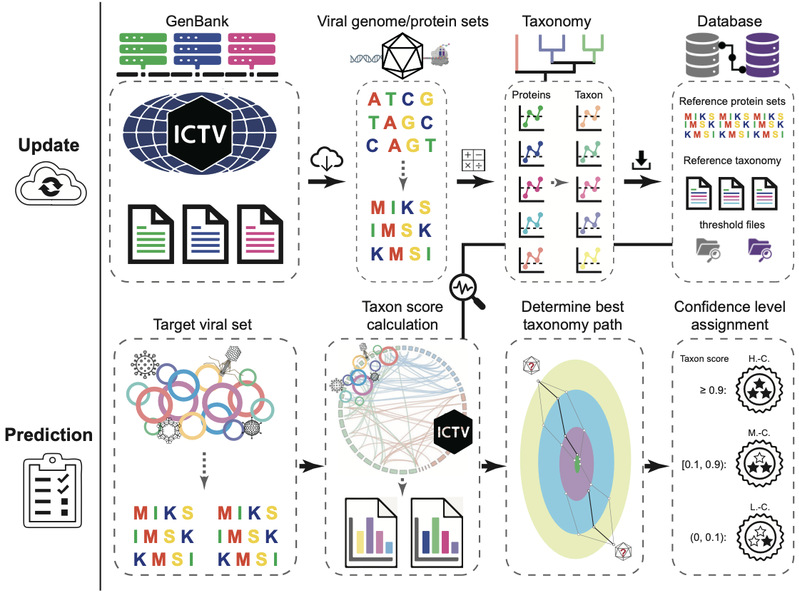
Figure 1. Flowchart of VITAP workflow and methodology.
With the rapid advancement of big data models, the exponential growth of data presents both opportunities and challenges for scientific research. Problems such as large data scale, structural heterogeneity, and inefficiencies in analysis have rendered traditional data analysis and classification methods increasingly inadequate to meet current scientific research needs. The accuracy of viral classification tools has long been a bottleneck in advancing research efficiency. To address this problem, Professor Wang and her team conducted systematic comparative analyses of existing classification tools within the virology domain. While ensuring high accuracy, VITAP achieved more comprehensive and stable viral classification, providing an efficient automated tool for studies in viral genomics, taxonomy, and ecology. At the initial research stage, the team drew inspiration from classification tools in different domains to propose preliminary algorithm designs. However, the scientific journey encountered various challenges from framework design to manuscript submission. Confronting these difficulties, the researchers persevered through extensive literature review, countless validations and revisions, ultimately overcoming the obstacles and successfully publishing their work.
Article link:
https://www.nature.com/articles/s41467-025-57500-7



